

| Home | News | Download | Features | Documentation | FAQ | Installation | Feedback |
 Concoord version 2.1.2 , a minor bugfix and performance increase release has been released.
Concoord version 2.1.2 , a minor bugfix and performance increase release has been released.
Concoord version 2.1 is ready! Among the new features are:
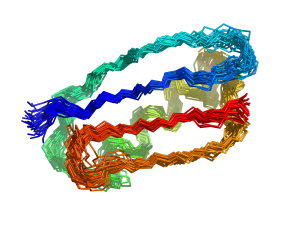
CONCOORD is a method to generate protein conformations around a known structure based on geometric restrictions. Principal component analyses of Molecular Dynamics (MD) simulations of proteins have indicated that collective degrees of freedom dominate protein conformational fluctuations. These large-scale collective motions have been shown essential to protein function in a number of cases. The notion that internal constraints and other configurational barriers restrict protein dynamics to a limited number of collective degrees of freedom has led to the design of the CONCOORD method to predict these modes without doing explicit, more CPU intensive, MD simulations.
The CONCOORD method consists of two stages. The first stage is the identification of all interatomic interactions in the starting structure. These interactions are divided among different categories, depending on the strength of the interaction. Covalent bonds form the tightest interactions and long-range non-bonded interactions are among the weakest interactions. Based on the strength of the interaction, a specific geometric freedom is given to each interacting pair. In this way, a set of upper and lower geometric bounds is obtained for all interacting pairs of atoms. The second stage consists of generating structures other than the starting structure that fulfill all geometric bounds. This is achieved by iteratively applying corrections to randomly generated coordinates untill all bounds are fulfilled.
The CONCOORD software consists of two programs, one for each stage. The program dist generates the geometric bounds and disco is the program that generates structures from these bounds. Use the -h option of both programs for detailed usage information. These two programs will be put in the PATH environment variable by sourcing the CONCOORDRC file in the CONCOORD root directory. Additionally, the location of a number of library files will from then on also be known to the programs. An interface to CONCOORD can be found in the WHAT IF and YASARA packages. Additionally, Dynamite is based on CONCOORD.
Concoord is free to use for everyone. However, if you use concoord for publications or presentations you must cite the original concoord publication:
B.L. de Groot, D.M.F. van Aalten, R.M. Scheek, A. Amadei, G. Vriend and H.J.C. Berendsen; "Prediction of protein conformational freedom from distance constraints", Proteins 29: 240-251 (1997) [pdf]
The previous distributions are also still available:
(C) Bert de Groot, 1996-2010
Please note that the software is distributed with NO WARRANTY OF ANY KIND. The author is not responsible for any losses or damages suffered directly or indirectly from the use of the software. Use it at your own risk.
Please send your bug reports, comments and suggestions to:
bgroot@gwdg.de.
Enjoy!