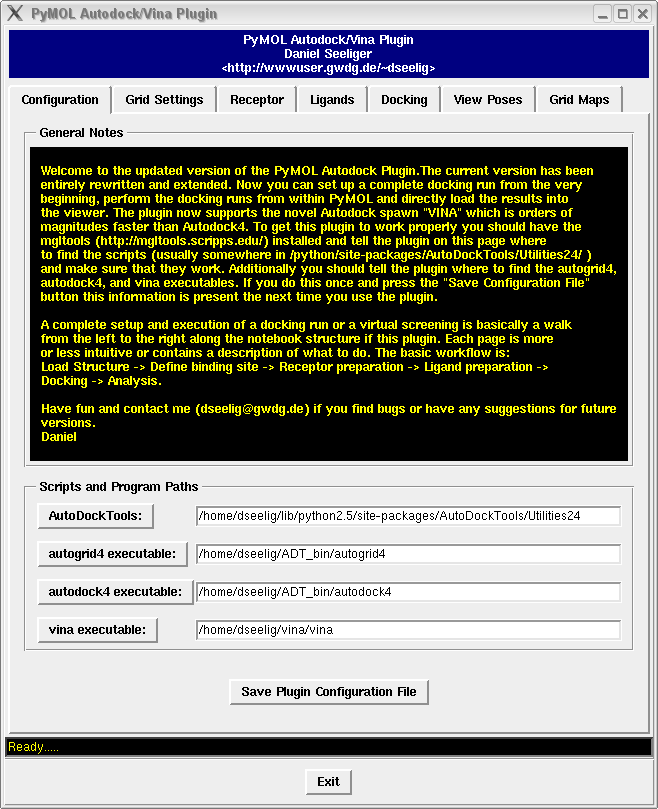
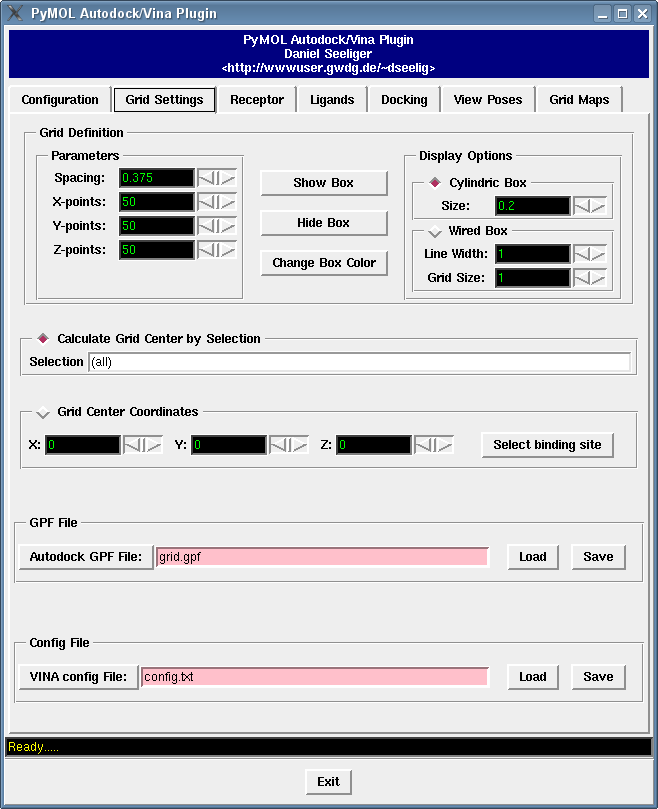
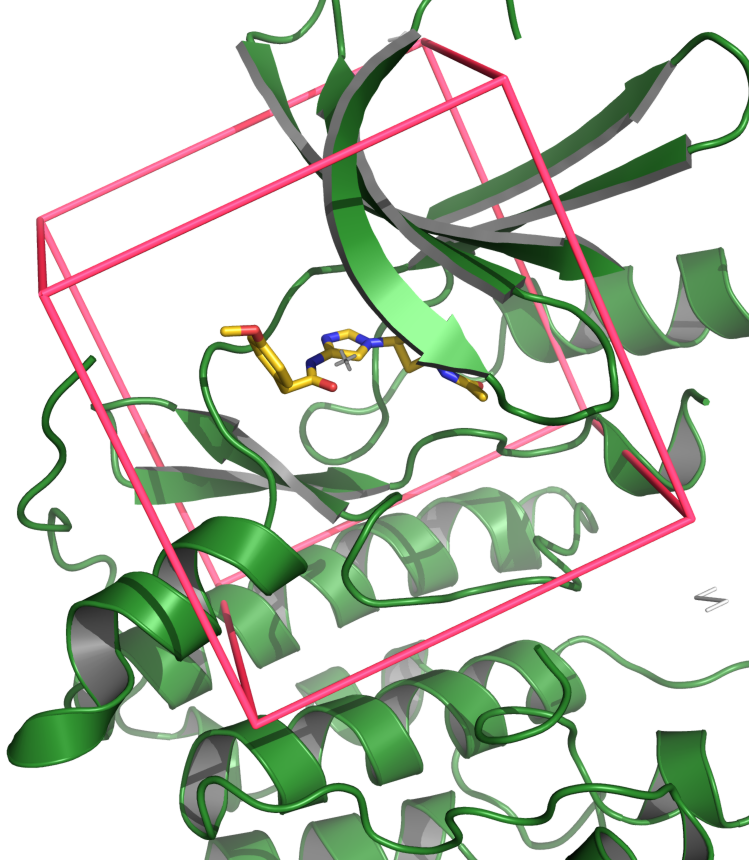
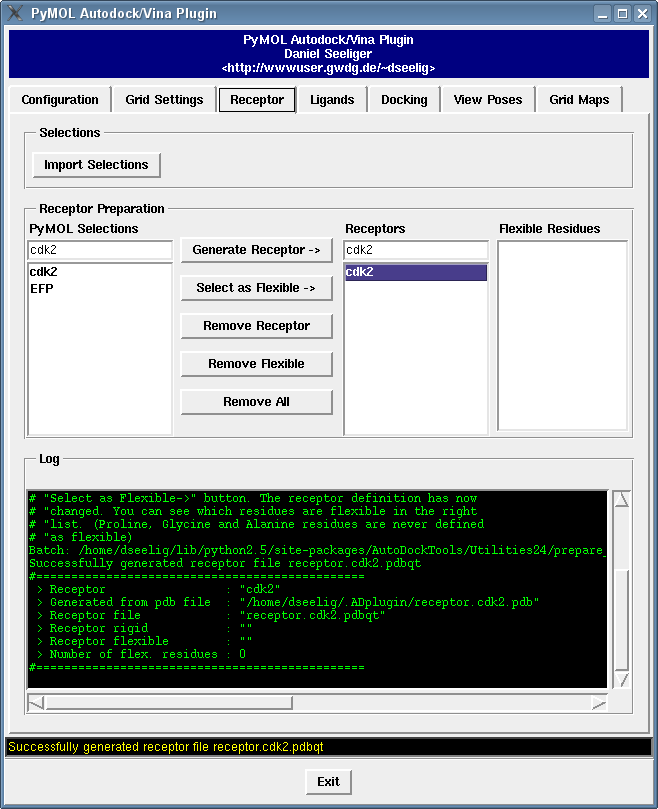
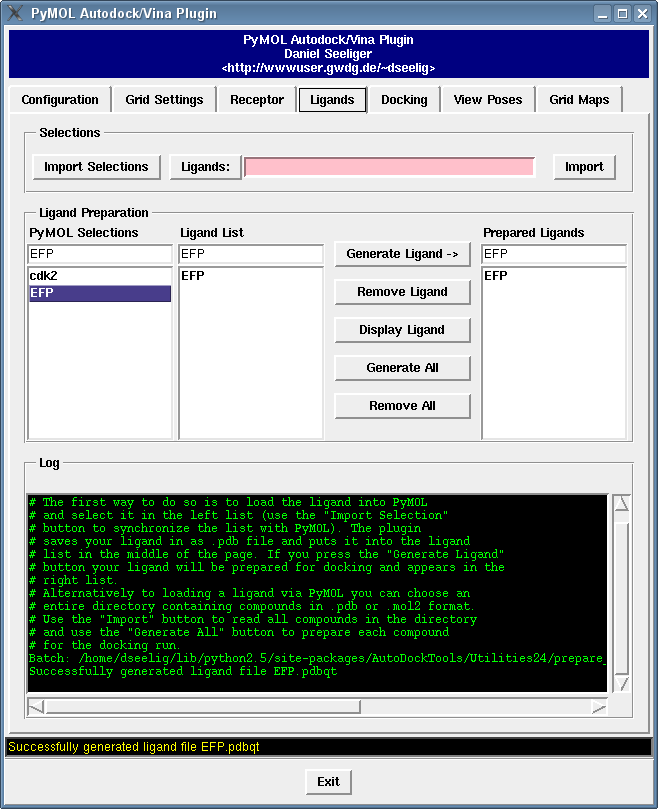
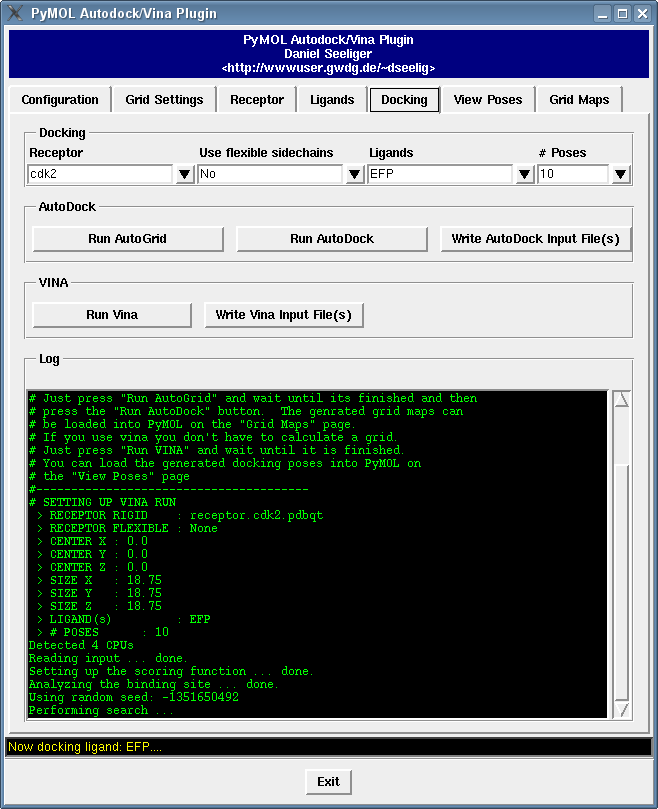
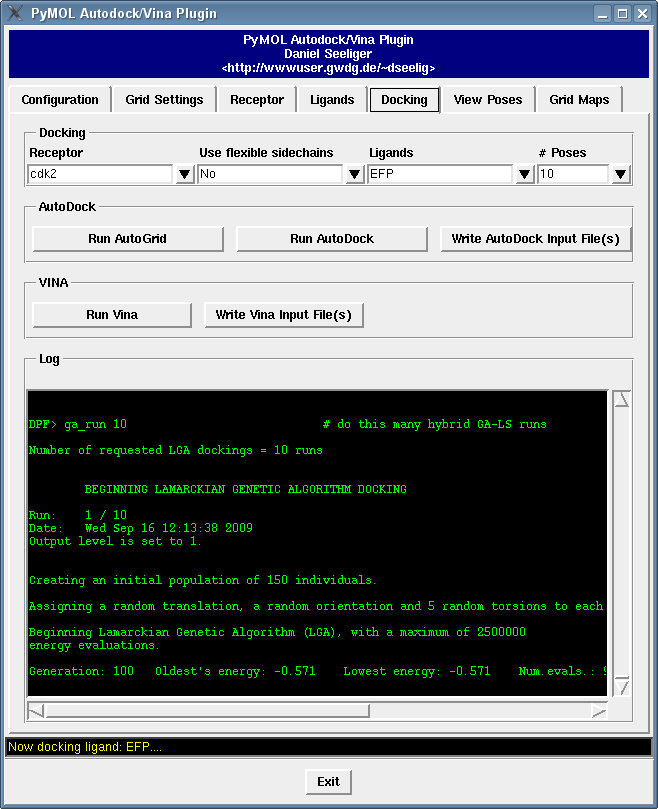
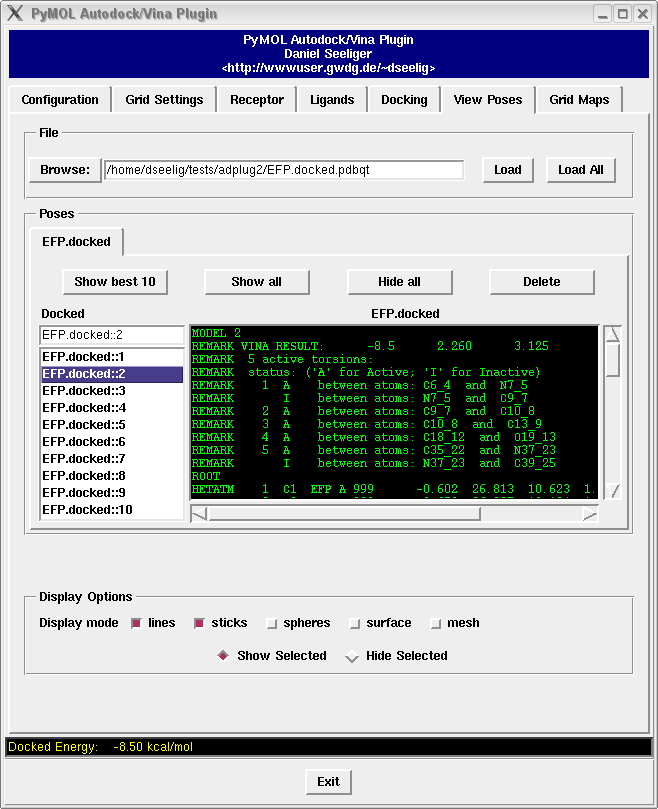
It contains a bunch of new features such as
learn how to set up a virtual screening with the virtual screening tutorial,
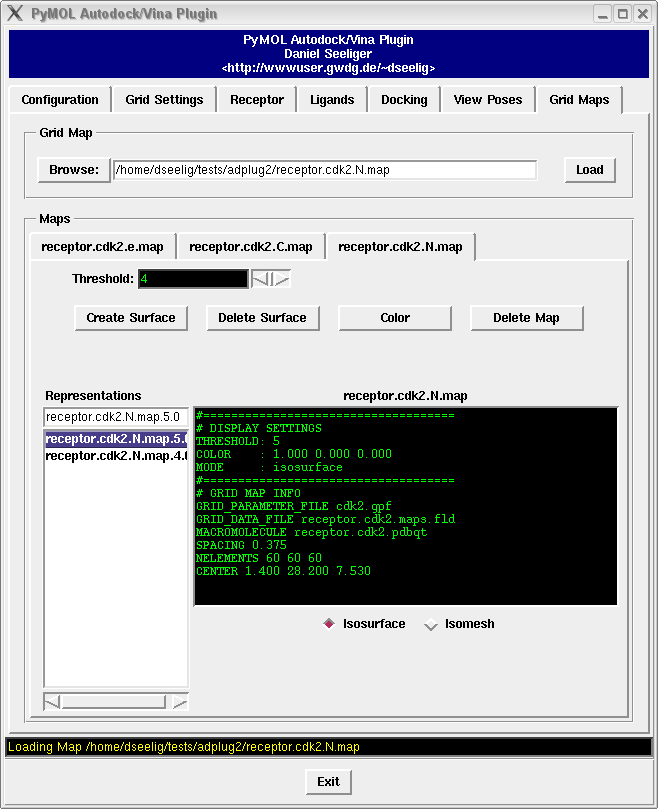
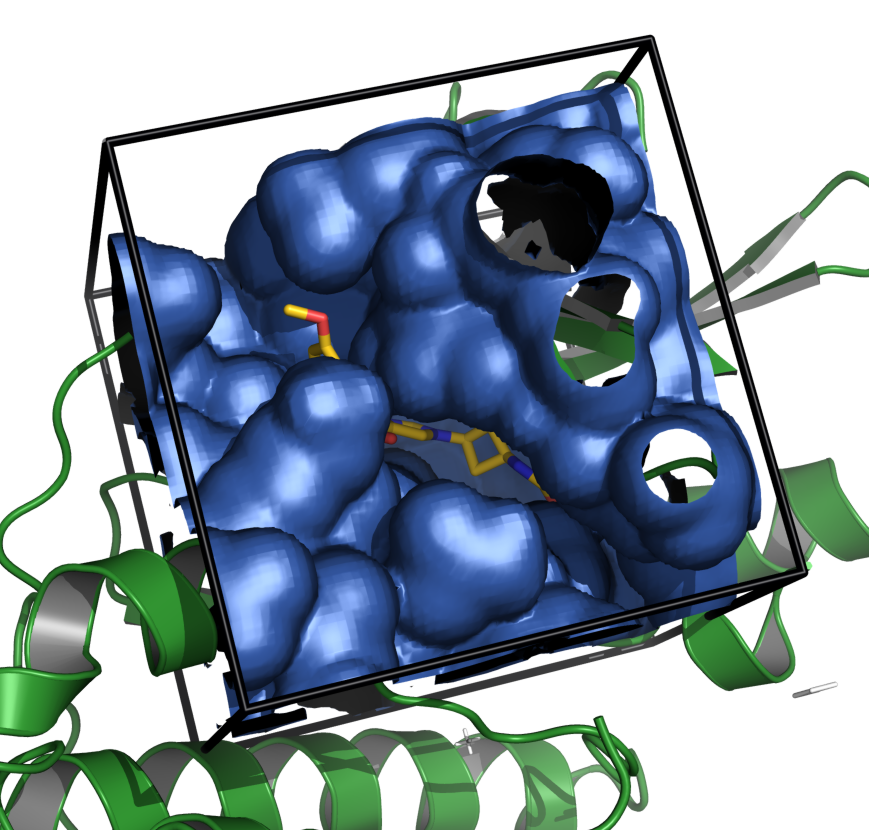
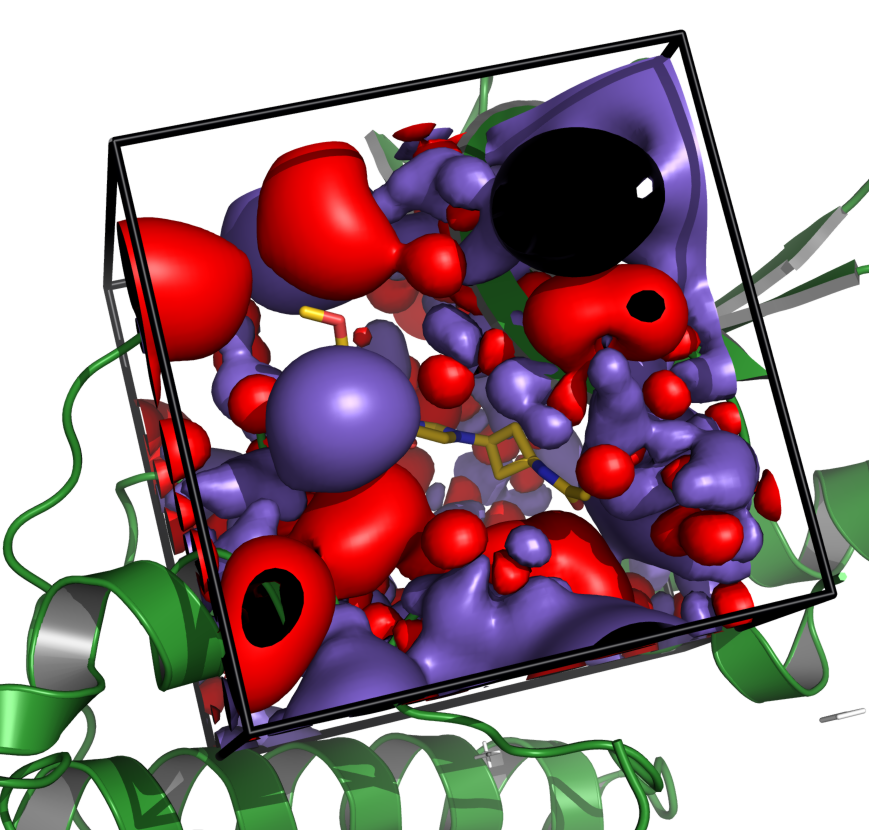
see how you can deal with grid maps with the grid map tutorial,
and get an idea about docking runs with flexible sidechain with the flexible sidechains tutorial.
Download plugin here: https://github.com/ADplugin/ADplugin
SOME NOTES:
The plugin is developed and tested on Linux. I have no other possibilities to test it and to fix bugs related to other operating systems. However, if you use the plugin on a Mac and experience some errors when scripts from the MGLTools package are executed, try to replace the first line in these scripts.
#!/usr/bin/env pythonshould be replaced by
#!/usr/bin/env /Library/MGLTools/1.5.4/bin/pythonsh( note the whitespace between env and /Library )
The scripts executed by the plugin are:
I sometimes experienced crashes (segmentation faults) when executing autogrid or autodock. This is due to redirecting the log file output to the plugin text viewer window. You can switch this feature off by setting the environmental variable "ADPLUGIN_NO_OUTPUT_REDIRECT".
Ligand names should not contain an underscore, e.g. ligand_1.pdb, rename it to ligand-1.pdb.
Happy Docking!