:
:

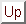

To enable studies of low-frequency protein dynamics by extended MD-simulations of a simplified protein model, the design of that model has to meet two main requirements: (a) the model should exhibit structural and dynamical properties similar to those of real proteins, in particular, it should enable a study of conformational transitions; (b) the model should exhibit as few degrees of freedom as possible, and, accordingly, should be structurally simple. Thus, the protein model should represent a `minimal' model. Such model can not contain all features of proteins, nor can it model the dynamics of a specific protein. Therefore, we included only those structural elements, the combination of which a priori seemed to be essential for low-frequency dynamical properties.
For the design of the protein model we have chosen a two step procedure: first, we defined a `primary' structure, consisting of 100 residues, and a force field. Second, we simulated a `folding' process of that model in order to obtain a stable tertiary structure.
To optimally meet the above conflicting objectives, we decided
to neglect the internal structure of the residues and to describe the
polypeptide by a chain of 100 van der Waals spheres,
which are linearly connected via interactions resembling chemical bonds.
The employed force field included
bond stretch, bond angle, van der Waals
and Coulomb interactions :
:

where the energy contributions are defined as in Ref. Brooks83.
The particle masses and force parameters were those of
 `extended atoms' and associated single bonds, as
defined in the CHARMm force field[23].
`extended atoms' and associated single bonds, as
defined in the CHARMm force field[23].
With the above definitions, the model describes a 100-alkane (`hektane')
rather than a protein. Therefore, additional properties which can mimic
the low-frequency behavior of proteins have to be
included. In order to identify such properties we note that
a characteristic feature of proteins is their ability to fold into
and to maintain a unique tertiary structure in native
environment[59]. That property is
a prerequisite for their specific biochemical function.
The tertiary structure is determined by
specific interactions of particular amino acid side groups,
e.g. by disulfide bonds or by H-bonds as well as by less specific
long-range interactions like
Coulomb or hydrophilic and hydrophobic interactions.
The latter type of interaction, in particular the hydrophobic force,
is known to dominantly contribute to the stability
of folded proteins[60,61].
We therefore decided to add heterogeneous
long-range interactions to our protein model.
in native
environment[59]. That property is
a prerequisite for their specific biochemical function.
The tertiary structure is determined by
specific interactions of particular amino acid side groups,
e.g. by disulfide bonds or by H-bonds as well as by less specific
long-range interactions like
Coulomb or hydrophilic and hydrophobic interactions.
The latter type of interaction, in particular the hydrophobic force,
is known to dominantly contribute to the stability
of folded proteins[60,61].
We therefore decided to add heterogeneous
long-range interactions to our protein model.
Primary structure:
For that purpose
we defined an artificial, heterogeneous `primary structure' by
assigning different partial charges to the 100 van der Waals spheres
of the model. Figure  shows
the chosen charge distribution
(bold, wavy curve) along the stretched chain.
As can be seen, that distribution divides the protein
model into five parts, three of which carry a positive charge, while the
remaining two are charged negatively. The inset of Fig.
shows
the chosen charge distribution
(bold, wavy curve) along the stretched chain.
As can be seen, that distribution divides the protein
model into five parts, three of which carry a positive charge, while the
remaining two are charged negatively. The inset of Fig.  shows the detailed structure of the model.
shows the detailed structure of the model.

Figure: Protein model in a stretched, unfolded configuration,
consisting of 100  -like `atoms'; their
partial charges are represented by the bold curve;
the inset shows a zoom of the detailed structure.
-like `atoms'; their
partial charges are represented by the bold curve;
the inset shows a zoom of the detailed structure.
The average absolute charge was chosen such that
the corresponding Coulomb energy contribution becomes sufficiently large as to
over-compensate entropic free energy contributions at room
temperature and thus to stabilize folded structures.
and thus to stabilize folded structures.
In that respect, the chosen
Coulomb interactions mimic hydrophobic protein-solvent
interactions comparable to those included into the bead-model
in Ref. Honeycutt92:
the attraction of oppositely charged `residues' resembles
hydrophobic forces, whereas the repulsion of equally charged `residues'
models the tendency of hydrophilic groups to solvate.
We note, that, as a consequence of the above interpretation, the protein
model comprises a simple effective solvent model.

Figure: Snapshots of the protein structure during the
simulated folding process described in
the text; structures are shown as `ribbon-plots' (a)--(f): initial,
completely unfolded
configuration (a); successive configurations after 3 ps (b), 6 ps (c),
7 ps (d), 8 ps (e), and 100 ps (f), respectively;
(g): folded structure of the protein model after 5 ns,
which was used as initial
configuration for the dynamics simulations described in the text;
the bold lines represent chemical bonds; four numbered circles mark atoms
referred to in the text.
Folding Process:
The second step --- the `folding' of the protein model --- was carried out by means of MD-simulation. All simulations presented in this paper have been performed in vacuo using the Verlet-algorithm[36] with an integration step size of one femtosecond for the integration of the Newtonian equations of motion. No `cut-off' has been employed.
Starting from an unfolded configuration, shown in
Fig.  and, as a ribbon-plot, in Fig.
and, as a ribbon-plot, in Fig.  (a),
the protein model was allowed to freely move under the influence
of bond-, van der Waals-, and Coulomb interactions.
Figure
(a),
the protein model was allowed to freely move under the influence
of bond-, van der Waals-, and Coulomb interactions.
Figure  (b--f) shows snapshots of the structure during
this initial phase of the `folding process'.
As can be seen, the compactness of the model rapidly increased
and, after 10 picoseconds, came close to its final value.
At this stage, the tertiary structure had not
yet stabilized, and the model exhibited frequent conformational changes.
(b--f) shows snapshots of the structure during
this initial phase of the `folding process'.
As can be seen, the compactness of the model rapidly increased
and, after 10 picoseconds, came close to its final value.
At this stage, the tertiary structure had not
yet stabilized, and the model exhibited frequent conformational changes.
Within the first few femtoseconds of the folding process,
the temperature of the model raised from 300 K to above 10,000 K.
This large temperature jump is due to high conformational energy
present in the initial structure, part of which
quickly converted into kinetic energy.
We continued this high
temperature dynamics for two nanoseconds to
explore configurational space.
By rescaling of atomic velocities,
the model was then slowly cooled down to 300 K, where it was
trapped in a well-defined
conformation, which remained stable during
the subsequent equilibration phase of one nanosecond duration.
The model was then allowed to move freely again for two nanoseconds
in order
to approach thermal equilibrium. This has been achieved, as is indicated
by the fact that no temperature drift could be observed during that
period.
The resulting, folded and relaxed conformation is
depicted in Fig.  (g). This structure was
used as initial configuration for the simulations described in
the following Sections.
(g). This structure was
used as initial configuration for the simulations described in
the following Sections.