Current Work
-
Processivity Mechanism in Sugar Degradation by Hyaluronate Lyase
- Since January 2004, I am working towards my PhD in Dr Bert de Groot's group at Max Planck Institute for Biophysical Chemistry, Goettingen, Germany. The main motivation behind the project is to study at atmoic level, the processivity mechanism of a prototypic enzyme called Hyaluronate Lyase. Processive enzymes differ from the normal enzymes in the property that that they remain attached to their polymeric substrates between the rounds of catalysis, and thus increase the efficiency manyfold. Being an inherently dynamic process, the mechanism of processivity, particularly the atomistic details of the substrate sliding phase between subsequent rounds of catalysis are difficult to be explored by exeperimental methods. We are trying to solve the problem by running many multi-nanosecond Molecular Dynamics (MD) simulations, both free and enforced type. A particular focus is on the question of how the enzyme can provide on one hand strong, specific binding during catalysis and on the other hand weak, unspecific binding during the substrate sliding phase.
- Currently, we have taken Streptococcus Pneumoniae Hyaluronate Lyase (Spn. HL) as a prototypic processive enzyme for this study. Spn. HL is a surface enzyme of Gram-positive bacterium, and it degrades hyaluronan and chondroitin/chondritin sulphates by cleaving the beta-1,4-glycosidic linkage between the glycan units of these polymeric substrates. This degradation makes the way for the bactrium through the host tissue thus spreading the disease process caused by pneumococci. Out of the two polymers, degradation of Hyaluronan alone is thought to proceed through a processive mechanism. The high-resolution structures of complexes between the enzyme both apo and holo (in complex with Hyaluronan as the substrate) [ref] have allowed for the structural insights into the degradation process of this polymeric substrate. However, the substrate translocation mechanism between two catalytic rounds is still unknown.
-
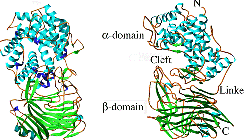
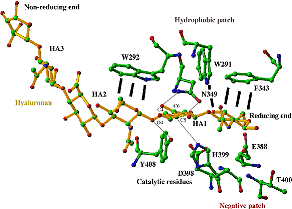
Figure 1: The structure of Spn HL showing two domains alpha and beta joined with a hinge loop. Figure 2: The proposed mechanism of Hyaluronan degradation. - With the help of sub-microsecond MD simulations of free and in complex with the hyaluronan (hexasaccharide) [PDB code: 1LOH] it has been elucidated that apo HL has more flexibility than the HL with the hyaluronan substrates present. This flexibility of the protein explored through the Principal Component Analysis (PCA) has independently predicted the new structures recently resolved experimentally.
-
The flexibility analysis, suggests the first three eigenvectors may also be involved in the processivity mechanism of HA degradation. These three eigenvectors are as follows:
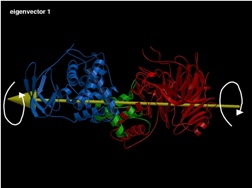
eigenvector 1: Twisting of the two structural domains (alpha and beta) through the hinge-loop.
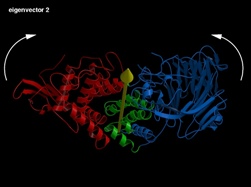
eigenvector 2: Opening/Closing of the catalytic cleft region
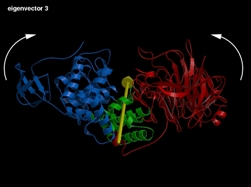
eigenvector 3: Opening/Closure of the access/exit of the catalytic cleft. -
Fig 3 (above) : The projection of apo and holo simulations on the first two principal components showing free HL is more flexible opening to the areas not explored before by the experiments. The simulations start from "Closed" conformation [PDB code: 1LOH] and the apo HL trajectory goes upto more "Open" conformation. The two red circles called PEGMME are the predicted structres also resolved experimentally recently.
Fig 4 (on the right) : The first three eigenvectors giving the largest structural motions of the protein.
- We are currently doing the forced simulations namely Forced Probe Molecular Dynamics (FPMD) and Essential Dynamics (ED) Simulations of different lengths of the hyaluronan substrates to see if there is any dependency of the length on the processivity mechanism. We have recently found that in the presence of longer substrates, product leaves spontaneuously, suggesting a possible minimum length dependence on the processive mechanism. We are currently incvestigating, the details of this dependence of this length in detail.